Enrichment and characterisation of methane-oxidising bacteria from
the estuarine environment.
6.1 Introduction
A group
of methylotrophs known as methanotrophs, are bacteria capable of utilising
methane as their sole source of carbon and energy (Anthony, 1975,1982).† These methanotrophic bacteria have generated
much interest due to their important role in carbon cycling (Hanson, 1980; Rudd
and Taylor, 1988), the environmental importance of methane oxidation and
because of their potential industrial and biotechnological applications
(Higgins et al., 1980; Cicerone and
Oremland, 1988; Hanson et al., 1991;
King, 1992; Oldenhuis and Jannsen, 1993; Reeburgh et al., 1993). An
account of methanotrophy in general is discussed in Chapter 1.
6.1.1 Isolation of
methanotrophs
The
existence of methane-oxidising bacteria has been known for many years, but
attempts to isolate them in pure culture have generally been unsuccessful.† The first isolation of a methane-oxidising
bacterium was from leaves and stems of the aquatic macrophyte Elodea (Sohgen, 1906) which was named Bacillus methanicus. In 1949 Hutton and
Zobell succeeded in isolating pure cultures of methane-oxidising bacteria from
several sources, including gas field soils, beach sand, and mud from marine and
freshwater samples.† An organism, which
appeared to be identical to Bacillus
methanicus, was isolated by Dworkin and Foster (1956) and called Pseudomonas methanica.† During the 1960s, several other
methanotrophs were isolated including Methanomonas
methanooxidans (Brown et al.,
1964), Pseudomonas methanitrican
(Davies et al., 1964) and Methylococcus capsulatus (Foster and
Davies in 1966).† In 1970 Whittenbury
and colleagues devised enrichments and isolation techniques that led to the
establishment of over 100 pure cultures of gram negative, strictly aerobic,
methane utilising bacteria being isolated from ponds, rivers, streams and
ditches.† From these cultures a basic
scheme of classification was evolved (Davis and Whittenbury, 1970; Whittenbury et al., 1970; Whittenbury and Dalton,
1981).† The classification of
methane-oxidising bacteria is discussed in Chapter 1.† Briefly, the classification of methanotrophs can be divided into
two groups, Type I and Type II.† Type I
methanotrophs possess bundles of intracytoplamic membranes (which are
restricted to a few photosynthetic bacteria (Pfenning, 1967) and nitrifying
bacteria (Murray and Wilson, 1969), use the RuMP pathway for carbon
assimilation into biomass.† These organisms
belong to bacteria in the g-subdivision
of the protobacteria.† Type II
methanotrophs, have unusual intracytoplasmic membranes arranged around the
periphery of the cell, utilise the serine pathway for carbon assimilation and
belong to the a-subdivision of the
protobacteria.† Their taxonomy has been
extensively reviewed by Green, 1992 and Bowman et al., 1993, 1995.
Since
1970 there have been other reports of isolation of methane-oxidising bacteria
from a variety of environments (e.g. DeBont et
al., 1978; Hanson and Wattenberg, 1991; Bowman et al., 1993;† Gal'chenko,
1994; Khmelenina et al., 1996;
Omelíchenko et al., 1996;† Bowman et
al., 1997).† However, isolation of
marine strains has been limited.†
Enrichment and isolation of Type I and Type II marine methanotrophs was
reported by Heyer et al., (1984) but
no characteristics of these organisms were given. In 1987 Sieburth and
co-workers enriched and isolated a methanotroph from the upper mixed layer of
the Sargasso Sea.† The bacterium
isolated from samples was a Type I methanotroph, Methylomonas pelagica.† This
strain has now been re-named Methylomicrobium
pelagicum.† Lidstrom (1988) isolated
four new methane-oxidising bacteria from the marine environment.† All of which required NaCl for growth and
had characteristics of both Type I and Type II methanotrophs.† Lees et
al., (1992) isolated 2 new methane-oxidising bacteria from seawater samples
and again these organisms had an obligate requirement for NaCl and exhibited
many properties of typical Type I methanotrophs.† Two marine methanotrophs have also been isolated from seawater
samples by Holmes et al., (1995),
called Methylomonas sp. IR1 and Methylomonas sp. DR1.† Finally, isolation of methanotrophs from
symbiotic relationships with deep sea mussels (Bathymodiolus) and pogonophora has occurred over the last decade
(Childress et al., 1986; Brocks et al., 1987; Cavanaugh et al., 1987; Wood and Kelly, 1989;
MacDonald et al., 1989; Schmaljohann et al., 1990; Lees et al., 1992; Distel and Cavanaugh, 1994).† Kochevar et
al., (1992) studied such symbiotic relationships, and showed that bacterial
endosymbionts displayed remarkable characteristics not found in their
free-living counterparts.
6.1.2 Molecular
characterisation of methane-oxidising bacteria
Microbial
ecology has long been hampered by the fact that many microorganisms cannot be
identified in situ because of the
lack of morphological diversity.†
Advances in microbial ecology (see Ward et al., 1992) make it possible to use molecular techniques to
overcome such problems. The obligate requirement of methanotrophs for methane
allows highly selective enrichment conditions to be established.† Furthermore, the construction of nucleic
acid probes for the detection and retrieval of specific sequences (Britschgi
and Giovannoni 1991; Schmidt et al.,
1991) is now a popular tool for elucidating the types of methane-oxidising
bacteria present in environmental samples and enrichment cultures.† These nucleic probes can be divided into two
groups, phylogenetic and functional gene probes (see Chapter 5).
Another
molecular technique, fluorescence in situ
hybridisation (FISH) has been used as a powerful tool in microbial ecology
(Amann et al., 1990a; 1995). The
principle of FISH relies on the specific annealing of a labelled nucleic acid
probe to complementary sequences in fixed bacterial sample, using fluorescent
or radiolabelled oligonucleotide, followed by visualisation of the location of
the probe (DeLong and Shah, 1990).
The
development of fluorescent labelled rRNA-targeted oligonucliotide probes
labelled with fluorescent dyes which span the entire phylogenetic spectrum from
domains to subspecies, has successfully been applied for the detection and
identification of individual microbial cells in situ (Giovannoni et al.,
1988; Stahl et al., 1989; Amann et al., 1992). This whole cell
hybridisation allows for quantification and identification of microbes in a
cultivation independent way, which makes it possible to define in detail the
occurrence, abundance and the distribution of microorganisms in the natural environment
(Amann et al., 1990; Cary and
Giovannoni, 1993; Erauso et al.,
1993; Manz et al., 1993; Wagner et al., 1993; Wagner et al., 1994; Reysenbach et al., 1994; Amann et al., 1995; Wagner et al.,
1995; Snaidr et al., 1997).†
Labelled
probes have been used to investigate microorganisms in a wide range of
environments including aquatic ecosystems (Hicks et al., 1992; Lim et al.,
1993), soil and sediments (Hahn et al.,
1992; DiChritina and Delong, 1993; Spring et
al., 1992) and activated sludge (Wagner et
al., 1993,1994).
6.1.3 Study aims
Methane
concentrations and oxidation rates have been determined from marine samples
(see Chapter 3).† These data have shown
the marine environment to be supersaturated with methane with respect to the
atmosphere.† Both methane fluxes and
oxidation rates in the water column and air sea interfaces suggest that
biological oxidation is an important factor in controlling the flux of methane
to the atmosphere and local coastal systems (Ward et al., 1987; Chapter 3).†
Therefore, it is important to understand and characterise the
methane-oxidising bacteria present in the marine environment.
The objective
of this study was to amalgamate traditional enrichment and isolation approaches
with molecular characterisation and thus to observe differences between methane
enrichments and detection of methane-oxidising bacteria from the estuarine environment.
The
specific aims of this study were:-
∑
To enrich for and attempt to isolate cultures of marine
methane-oxidising bacteria.
∑
To use the PCR amplification technique on enrichment
cultures with primers targeting the group specific sequences of 16S rDNA and
functional gene primers (encoding for sMMO and pMMO) specific for
methane-oxidising bacteria.†
∑
To explore the use of FISH to identify the types of
methane-oxidising bacteria present in enrichment cultures.†
6.2 Results
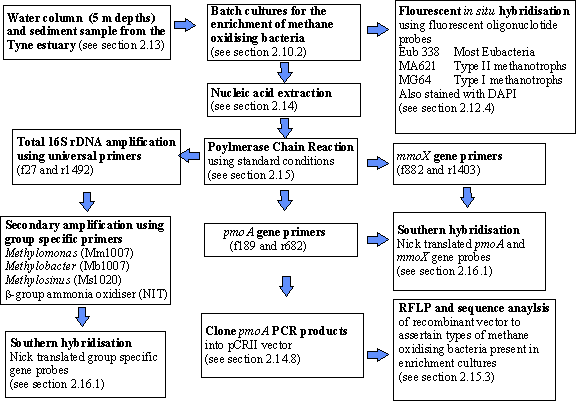
The
strategy of the protocols used in this study is shown in figure 6.0.† Enrichment culture isolation experiments
were performed with samples collected from the Tyne estuary at three different
stations (figure 6.1).
|
|
Figure 6.0 Flow chart to describe the protocols used to characterise
methane enrichments from the Tyne estuary.
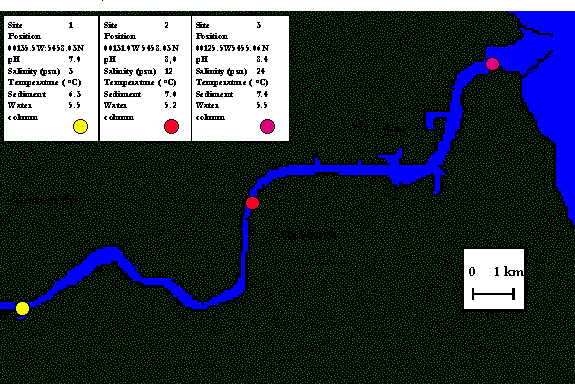
Figure 6.1 Description of
sampling sites and locations on the Tyne estuary
Samples
were obtained from the Tyne estuary as described in section 2.2 and were stored
at 4 oC until enrichment cultures were established with methane, as
the sole carbon source.
6.2.1 Characterisation of enrichments
Enrichment
culture techniques have classically been used to isolated bacteria capable of
utilising a particular substrate.† This
method is particularly useful for methane-oxidising bacteria and is based on
the selective growth advantage gained by an organism that is capable of using
methane as the sole carbon and energy source.†
Batch
enrichment cultures were set up from all estuarine samples (34 in total) by
direct amendment of seawater and sediment samples with basic mineral salts (see
section 2.10.2) using methane as the sole carbon source.† All enrichment cultures showed growth in 4-6
weeks. Due to the restriction of facilities, after a period of nine months ten
enrichments that showed greatest biological growth (assessed by measuring
optical density (540 nm)) were maintained for the period of the study.† It is possible that some enrichments were
more successful than others due to different populations of methane-oxidising
bacteria being capable of growing to higher cell densities.
Heterotrophs
were found to be present in all cultures, this was determined by light
microscopy, although morphology indicated that each enrichment culture was
dominated by a single heterotroph.† All
cultures were found to form clusters within liquid media, which is common among
bacterial enrichments from the marine environment (Mitchell et al., 1996).† However, before the cultures were discarded those which showed
some selection were harvested as described in section 2.7 using 25 ml sample
volume.† These preparations were used to
increase samples for molecular biological techniques.
Following
the liquid sub-culturing procedure described in section 2.10.2, it was
desirable to obtain discrete colonies on a solid surface medium.† Marine methanotrophic organisms are known to
be difficult to cultivate on agar-based media (Lees et al., 1991). From this work, experiments to isolate methanotrophs
in pure culture were unsuccessful.†
Numerous attempts to plate out these samples failed, as previously
experienced (Lees et al., 1991;
Holmes et al., 1995a) demonstrating
the problems associated with the isolation of these organisms.
6.2.2 PCR experiments on DNA
from enrichments
High
molecular mass DNA was readily extracted from all batch culture samples (see
section 2.14.5).† Extracted DNA were then
put through PCR reactions to determine the presence/absence of either known
methanotrophs or the type of methane monooxygenase.†
6.2.2.1 Group specific
The 16S
rDNA oligonucleotides group specific probes for Methylomonas, Methylomicrobium, Methylosinus, and b subclass ammonia-oxidising
bacteria were used in PCR to screen DNA samples for the presence of putative
methane-oxidising bacteria. However, work by Utaker and Nes (1998), has
suggested that the NIT primers used in this study may, under certain
conditions, amplifiy false positives (see Chapter 5).
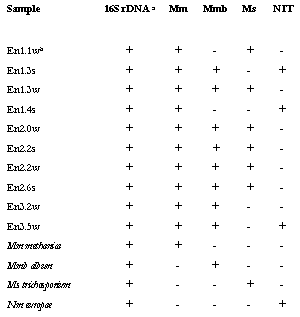
The
results of PCR experiments performed are shown in table 6.0. All methanotroph
group specific primers were used in combination with f27 and the relevant group
specific target region on the 16S rDNA, in a nested PCR reaction.† Thirteen PCR reactions failed to give a PCR
amplification product with group specific primers tested, but did give an
amplification product using universal eubacterial primers.† All primers gave a signal with the relevant
control organism.† Furthermore, it can
be seen that all the enrichments gave rise to PCR amplification products (of
the expected size) to at least one of the four group specific primers (table
6.0).† The most common (i.e. found)
methanotroph like sequence was of the genus Methylomonas.† Figure 6.2 demonstrates the PCR amplified
products of Methylomonas specific
primers were of the expected size and was observed in all enrichment samples
tested. These PCR products were not confirmed by Southern hybridisation due to
time constraint.† However, If the
products were to be confirmed by Southern hybridisation, a set of unique
primers would be needed (between f27 and r1492) that amplify only
methane-oxidising bacteria.† This is due
to the ability of a 16S rRNA gene probe of 1 kb in length to discriminate
between different genera of methylotrophs.†
A further experiment to test whether all four genera were present would
be to produce clone library of the 16S rDNA products.
Table 6.0 Description of 16S
rDNA PCR with eubacterial and group specific primers
|
|